This is an interface to simplify preprocessing of one, a subset or all
blocks in a multiblock object, e.g., a data.frame (see block.data.frame)
or list. Several standard preprocessing methods are supplied in addition to
letting the user supply it's own function.
Details
The fun parameter controls the type of preprocessing to be performed:
autoscale: centre and scale each feature/variable.
center: centre each feature/variable.
scale: scale each feature/variable.
SNV: Standard Normal Variate correction, i.e., centre and scale each sample across features/variables.
EMSC: Extended Multiplicative Signal Correction defaulting to basic EMSC (2nd order polynomials). Further parameters are sent to
EMSC::EMSC.Fro: Frobenius norm scaling of whole block.
FroSq: Squared Frobenius norm scaling of whole block (sum of squared values).
SingVal: Singular value scaling of whole block (first singular value).
User defined: If a function is supplied, this will be applied to chosen blocks. Preprocessing can be done for all blocks or a subset. It can also be done in a series of operations to combine preprocessing techniques.
See also
Overviews of available methods, multiblock, and methods organised by main structure: basic, unsupervised, asca, supervised and complex.
Common functions for computation and extraction of results and plotting are found in multiblock_results and multiblock_plots, respectively.
Examples
data(potato)
# Autoscale Chemical block
potato <- block.preprocess(potato, block = "Chemical", "autoscale")
# Apply SNV to NIR blocks
potato <- block.preprocess(potato, block = 3:4, "SNV")
# Centre Sensory block
potato <- block.preprocess(potato, block = "Sensory", "center")
# Scale all blocks to unit Frobenius norm
potato <- block.preprocess(potato, fun = "Fro")
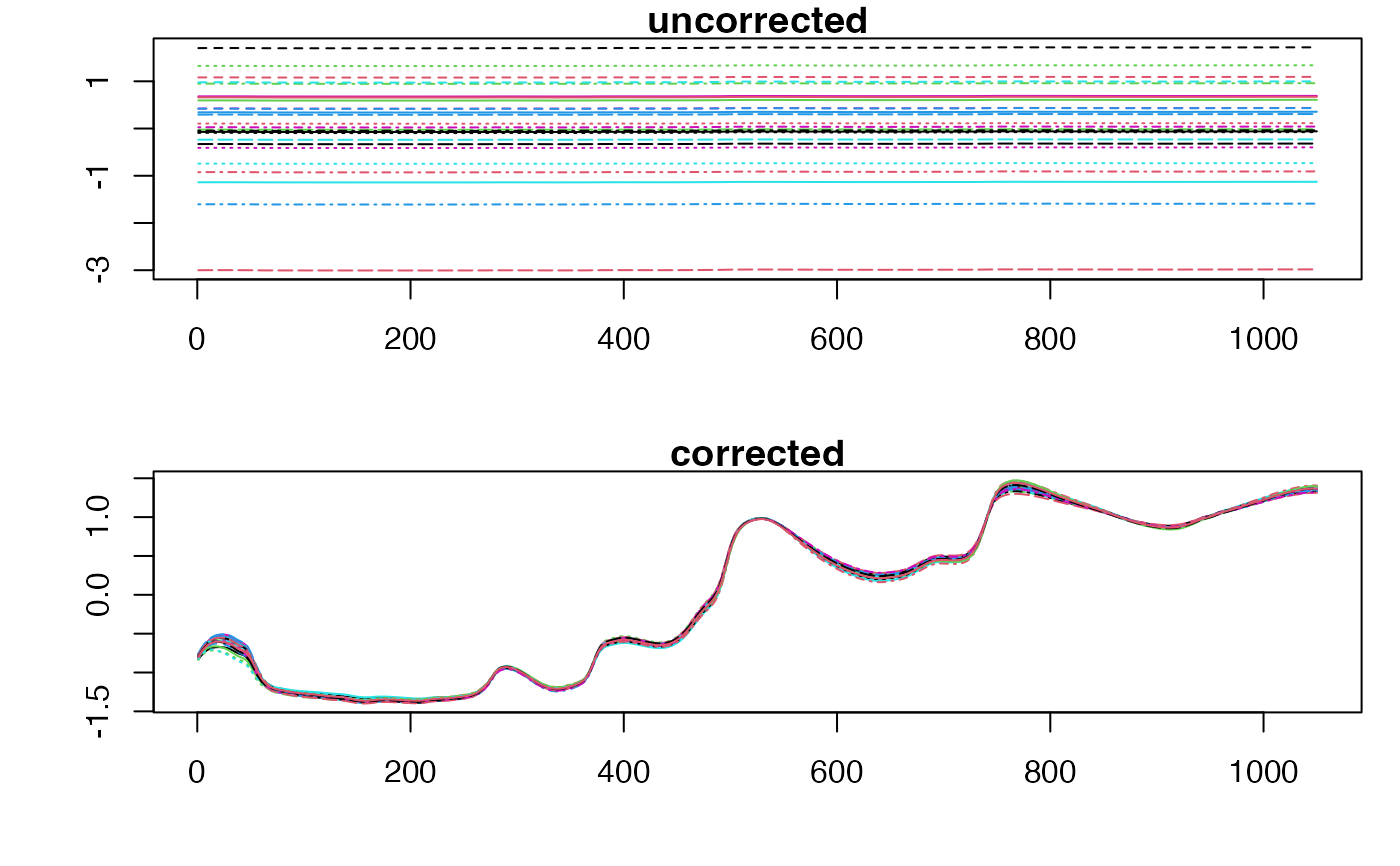
# Effect of SNV
NIR <- (potato$NIRraw + rnorm(26)) * rnorm(26,1,0.2)
NIRc <- block.preprocess(list(NIR), fun = "SNV")[[1]]
old.par <- par(mfrow = c(2,1), mar = c(4,4,1,1))
matplot(t(NIR), type="l", main = "uncorrected", ylab = "")
matplot(t(NIRc), type="l", main = "corrected", ylab = "")
 par(old.par)
par(old.par)