# Start the multiblock R package
library(multiblock)
#> Registered S3 method overwritten by 'plsVarSel':
#> method from
#> print.mvrVal pls
#> Registered S3 methods overwritten by 'multiblock':
#> method from
#> print.multiblock ade4
#> summary.multiblock ade4
#>
#> Attaching package: 'multiblock'
#> The following object is masked from 'package:stats':
#>
#> loadingsBasic methods
The following single- and two-block methods are available in the multiblock package (function names in parentheses):
- PCA - Principal Component Analysis (pca)
- PCR - Principal Component Regression (pcr)
- PLSR - Partial Least Squares Regression (plsr)
- CCA - Canonical Correlation Analysis (cca)
- IFA - Interbattery Factor Analysis (ifa)
- GSVD - Generalized SVD (gsvd)
The following sections will describe how to format your data for analysis and invoke all methods from the list above.
Prepare data
We use a selection of extracts from the potato data included in the package for the basic data analyses. The data set is stored as a named list of nine matrices with chemical, rheological, spectral and sensory measurements with measurements from 26 raw and cooked potatoes.
data(potato)
X <- potato$Chemical
y <- potato$Sensory[,1,drop=FALSE]Modelling
Since the basic methods cover both single block analysis, supervised and unsupervised analysis, the interfaces for the basic methods vary a bit. Supervised methods use the formula interface and the remaining methods take input as a single matrix or list of matrices. See vignettes for supervised and unsupervised analysis for details.
Common output elements across methods
Output from all methods include matrices called loadings, scores, blockLoadings and blockScores, or a suitable subset of these according the method used. An info list describes which types of (block) loadings/scores are in the output. There may be various extra elements in addition to the common elements, e.g. coefficients, weights etc. The names() and summary() functions below show all elements of the object and a summary based on the info list, respectively.
# PCA returns loadings and scores:
names(pot.pca)
#> [1] "loadings" "scores" "Xmeans" "explvar" "PCA" "info" "call"
summary(pot.pca)
#> Principal Component Analysis
#> ============================
#>
#> $scores: Scores (26x2)
#> $loadings: Loadings (14x2)
# GSVD returns block scores and common loadings:
names(pot.gsvd)
#> [1] "loadings" "blockScores" "GSVD" "info" "call"
summary(pot.gsvd)
#> Generalized Singular Value Decomposition
#> ========================================
#>
#> $loadings: Loadings (26x26)
#> $blockScores: Block scores:
#> - NIRraw (1050x1050), NIRcooked (1050x1050)Scores and loadings
Functions for accessing scores and loadings are based on functions from the pls package, but extended with a block parameter to allow extraction of common/global scores/loadings and their block counterparts. The default value for block is 0, corresponding to the common/global block. Block scores/loadings can be accessed by setting block to a number or name.
# Global scores plotted with object labels
scoreplot(pot.pca, labels = "names")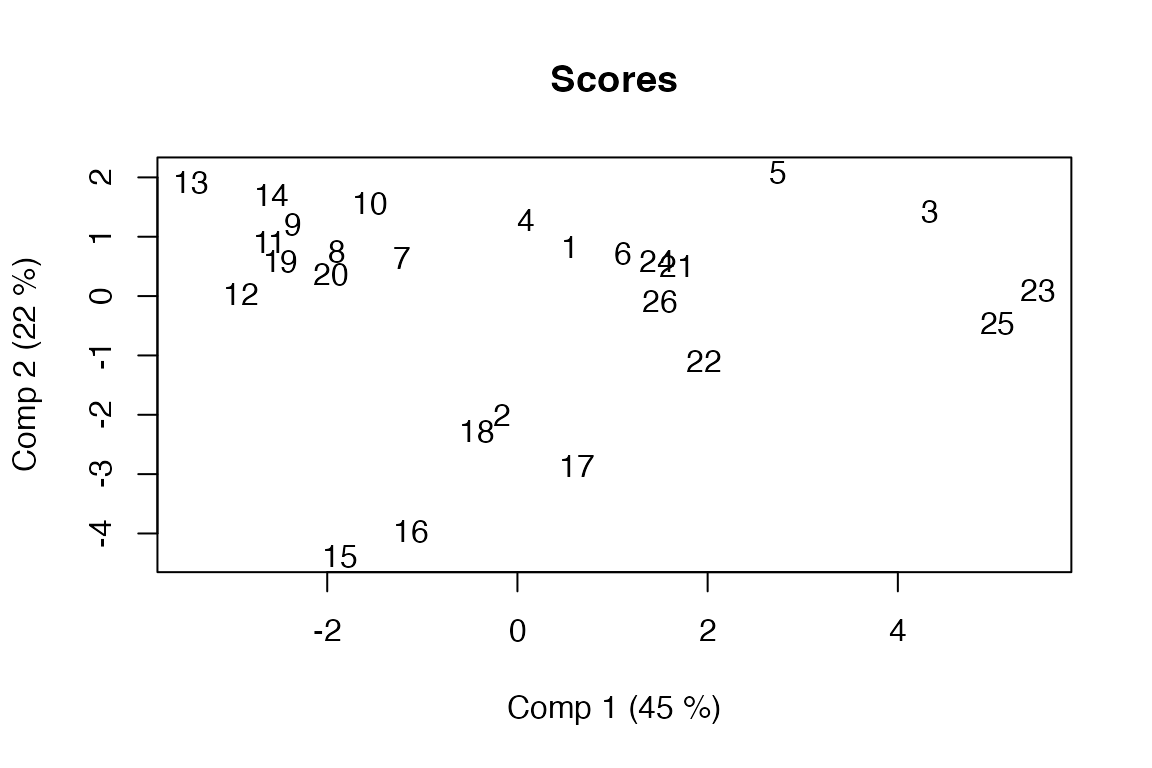
# Block loadings for Chemical block with variable labels in scatter format
loadingplot(pot.cca, block = "Chemical", labels = "names")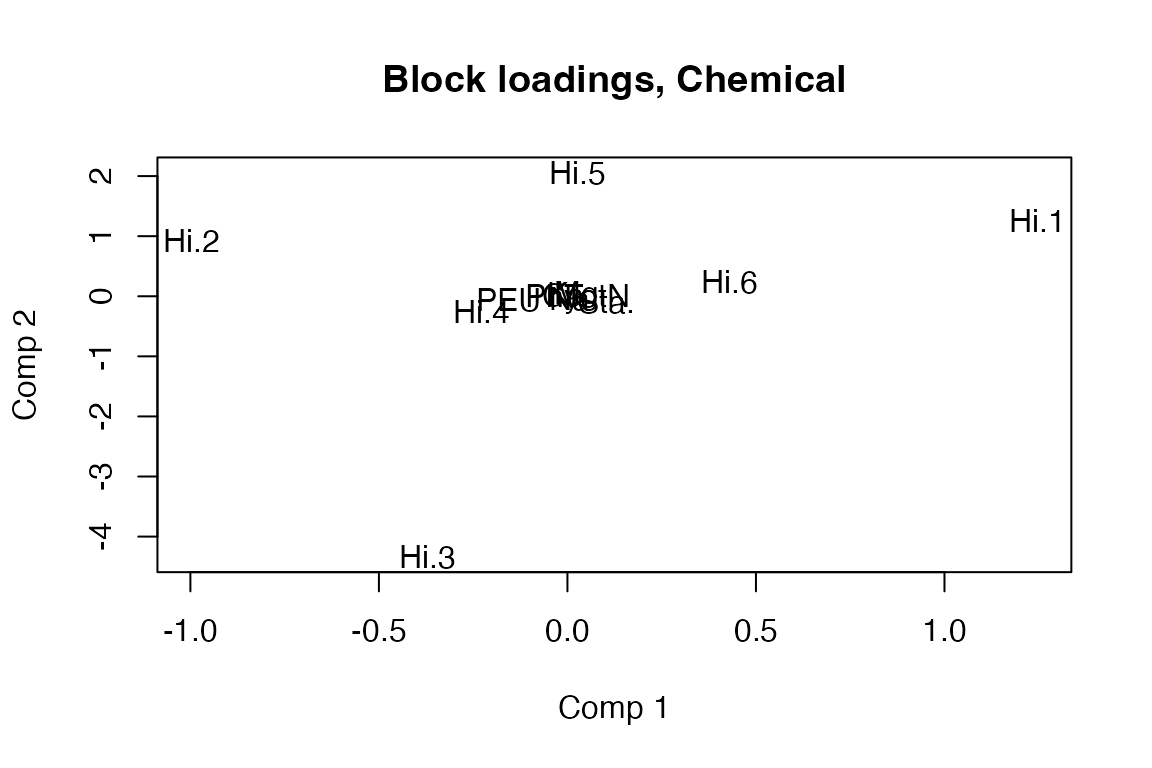
# Non-existing elements are swapped with existing ones with a warning.
sc <- scores(pot.cca)
#> Warning in scores.multiblock(pot.cca): No global/consensus scores. Returning
#> block 1 scores.