Fits a PLSR model with the SIMPLS algorithm.
Arguments
- X
a matrix of observations.
NAs andInfs are not allowed.- Y
a vector or matrix of responses.
NAs andInfs are not allowed.- ncomp
the number of components to be used in the modelling.
- center
logical, determines if the \(X\) and \(Y\) matrices are mean centered or not. Default is to perform mean centering.
- orthScores
logical. If
TRUEthe scores are orthogonalised for increased numerical precision (default = FALSE, for speed).- stripped
logical. If
TRUEthe calculations are stripped as much as possible for speed; this is meant for use with cross-validation or simulations when only the coefficients are needed. Defaults toFALSE.- ...
other arguments. Currently ignored.
Value
A list containing the following components is returned:
- coefficients
an array of regression coefficients for 1, ...,
ncompcomponents. The dimensions ofcoefficientsarec(nvar, npred, ncomp)withnvarthe number ofXvariables andnpredthe number of variables to be predicted inY.- scores
a matrix of scores.
- loadings
a matrix of loadings.
- Yscores
a matrix of Y-scores.
- Yloadings
a matrix of Y-loadings.
- projection
the projection matrix used to convert X to scores.
- Xmeans
a vector of means of the X variables.
- Ymeans
a vector of means of the Y variables.
- fitted.values
an array of fitted values. The dimensions of
fitted.valuesarec(nobj, npred, ncomp)withnobjthe number samples andnpredthe number of Y variables.- residuals
an array of regression residuals. It has the same dimensions as
fitted.values.- Xvar
a vector with the amount of X-variance explained by each component.
- Xtotvar
Total variance in
X.
If stripped is TRUE, only the components coefficients,
Xmeans and Ymeans are returned.
Details
This function should not be called directly, but through the generic
functions plsr or mvr with the argument
method="simpls". SIMPLS is much faster than the NIPALS algorithm,
especially when the number of X variables increases, but gives slightly
different results in the case of multivariate Y. SIMPLS truly maximises the
covariance criterion. According to de Jong, the standard PLS2 algorithms
lie closer to ordinary least-squares regression where a precise fit is
sought; SIMPLS lies closer to PCR with stable predictions.
References
de Jong, S. (1993) SIMPLS: an alternative approach to partial least squares regression. Chemometrics and Intelligent Laboratory Systems, 18, 251–263.
Examples
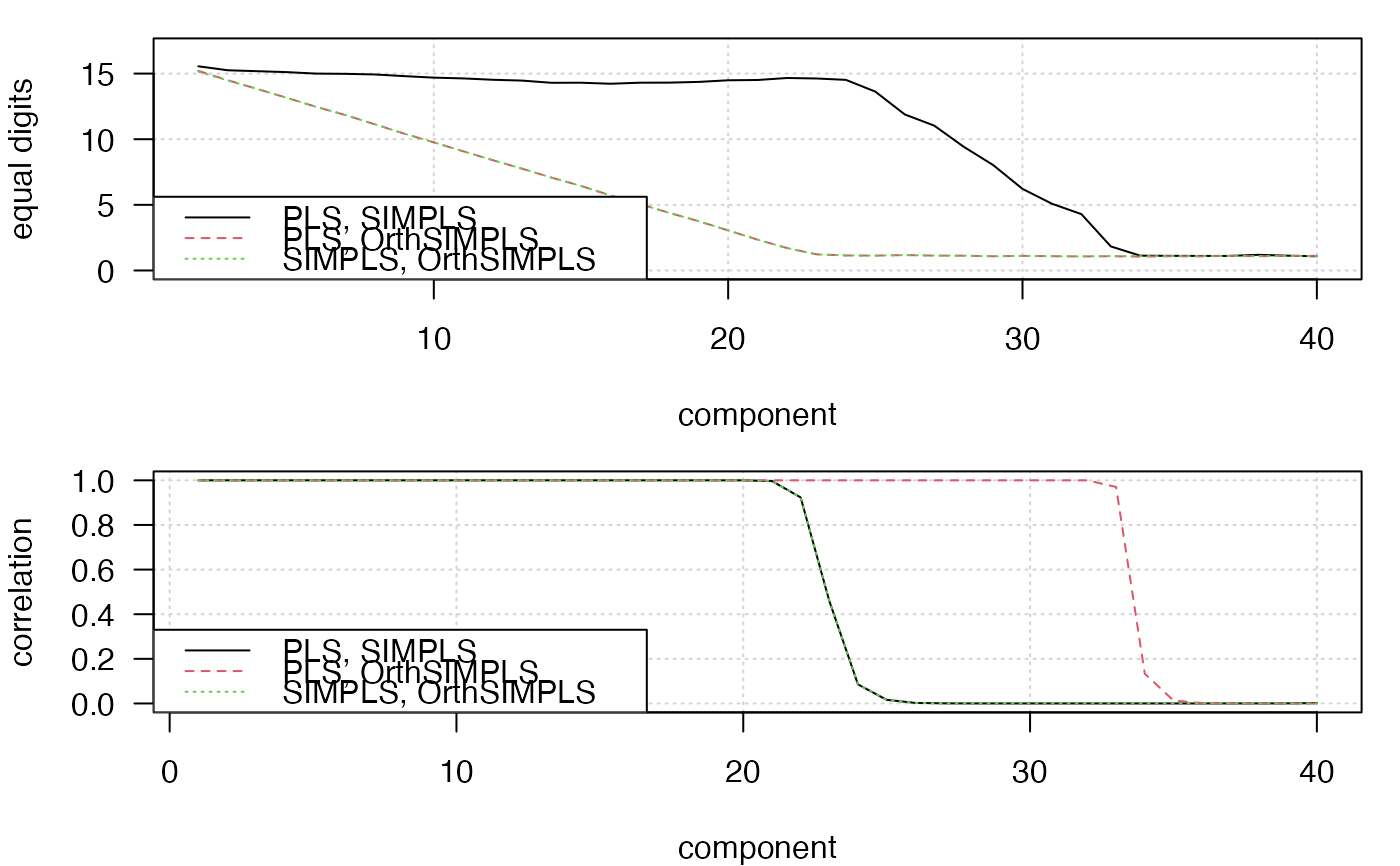
## Simulation of SIMPLS stability
# The graphics produced, demonstrate the numeric instability of the original
# SIMPLS without score orthogonalization.
set.seed(42)
N <- 100
p <- 2000
ncomp <- 40
simData <- data.frame(X = I(matrix(rnorm(N*p),N)), y = rnorm(N))
pls <- plsr(y ~ X, data=simData, ncomp=ncomp)
simps <- plsr(y ~ X, data=simData, ncomp=ncomp, method="simpls")
simpsO <- plsr(y ~ X, data=simData, ncomp=ncomp, method="simpls", orthScores=TRUE)
normScores <- pls$scores
for(i in 1:ncomp)
normScores[,i] <- normScores[,i]/sqrt(sum(normScores[,i]^2))
# Check number of equal digits
eqDig <- function(x,y){
xy <- abs(x - y)
xy[xy == 0] <- 10^-16
-colMeans(log10(xy))
}
eqDig_PLS_oSIMPLS <- eqDig(normScores, simpsO$scores)
eqDig_SIMPLS_PLS <- eqDig(simps$scores, normScores)
eqDig_SIMPLS_oSIMPLS <- eqDig(simps$scores, simpsO$scores)
# Correlation between models
cor_PLS_oSIMPLS <- diag(cor(pls$scores,simps$scores))
cor_SIMPLS_oSIMPLS <- diag(cor(pls$scores,simpsO$scores))
cor_SIMPLS_PLS <- diag(cor(simps$scores,simpsO$scores))
par.old <- par(mfrow=c(2,1), mar=c(4,4,1,1), las=1)
matplot(2:ncomp,cbind(eqDig_PLS_oSIMPLS,eqDig_SIMPLS_PLS, eqDig_SIMPLS_oSIMPLS)[-1,],
type="l", xlab="component", ylab="equal digits", ylim=c(0,17),
panel.first=grid())
legend("bottomleft", legend=c("PLS, SIMPLS", "PLS, OrthSIMPLS", "SIMPLS, OrthSIMPLS"),
col=1:3, lty=1:3)
matplot(1:ncomp,cbind(cor_PLS_oSIMPLS,cor_SIMPLS_oSIMPLS,cor_SIMPLS_PLS), type="l",
ylab="correlation", xlab="component", panel.first=grid())
legend("bottomleft", legend=c("PLS, SIMPLS", "PLS, OrthSIMPLS", "SIMPLS, OrthSIMPLS"),
col=1:3, lty=1:3)
 par(par.old)
par(par.old)